WHAT IS BIOPHYSICAL DYNAMICS?

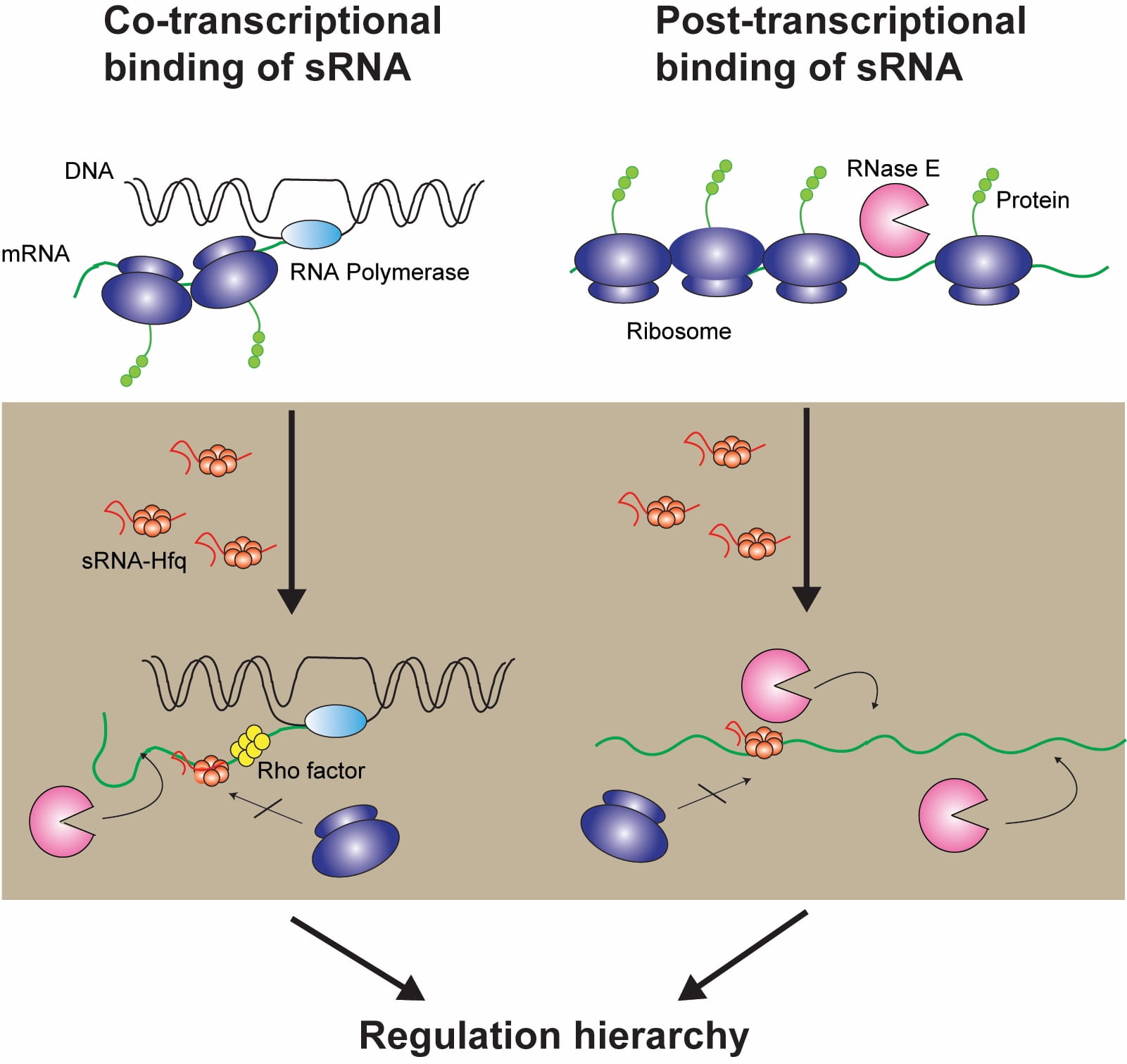
EVENTS
NEWS
Congratulations to Greg Engel on being named a co-Director of the Berggren Center for Quantum Biology and Medicine!
Congratulations to Benoit Roux on winning the 2024 Cole Award for excellence in research conducted in the field of membrane biophysics!
Congratulations to Margaret Gardel, Arvind Murugan and the entire team on being awarded $15.5 million by the NSF to establish the Center for Living Systems.
Congratulations to Greg Engel on being named an ACS Fellow!
Congratulations to Anthony Kossiakoff and Eduardo Perozo on being elected to the NAS!
Congratulations to Chuan He for winning the 2023 Wolf Prize for his pioneering work on RNA modifications!
Congratulations to Yamuna Krishnan on winning the NIH Pioneer Award!
Norbert Scherer named Optica C.E.K Mees Medal Winner. Congratulations Norbert!
Phoebe Rice has been named a Fellow of the American Association for the Advancement of Science. Congratulations, Phoebe!
The fundamental laws of physics and chemistry apply to all matter, yet living organisms can exploit these laws to create systems that can grow, adapt, process information, and ultimately self-replicate. How do these behaviors, the hallmarks of living systems, emerge from the interactions of non-living components? The Biophysical Dynamics Institute is home to a core group of researchers who work at the interface between the physical sciences and the biological sciences to answer these questions.
We are experimentalists, theoreticians, and computational scientists who have come together to address the challenge of obtaining a physical description of living systems. Our research spans scales from the study of molecules, which we seek to understand in terms of the interactions of atoms, to cells, which we seek to understand in terms of their molecular components, to biological networks, where connectivity and topology play defining roles. We are unified by the conviction that understanding dynamics, going beyond a static snapshot of equilibrium, will be key to future breakthroughs.
We support training programs and initiatives for undergraduates, graduate students, and postdoctoral fellows to advance interdisciplinary science at the University of Chicago with the goal of creating a culture of fluid exchange of ideas and collaboration across disciplines and among laboratories. These include close partnership with the Biophysics graduate program, and the Yen fellowship for outstanding postdocs in biophysics.
WHAT IS BIOPHYSICAL DYNAMICS?
We study the dynamics of
molecules, circuits, and cells